CLEAR item#35
“Training and tuning details. Describe the training process with adequate detail. Specify the augmentation technique, stopping criteria for training, hyperparameter tuning strategy (e.g., random, grid search, Bayesian), range of hyperparameter values used in tuning, optimization techniques, regularization parameters, and initialization of model parameters (e.g., random, transfer learning). If transfer learning is applied, clearly state which layers or parameters are frozen or affected.” [1] (from the article by Kocak et al.; licensed under CC BY 4.0)
Reporting examples for CLEAR item#35
Example#1. “For the classification task, a logistic regression ML classifier algorithm was selected based on specific characteristics of the dataset (expected number of instances available, use of polynomial transformation, tabular nature of the data) and used with a 5-fold stratified cross-validation (scoring metric = balanced accuracy) during the random search tuning process. The search space was defined as follows: penalty = l1, l2 or elasticnet, solver: saga or LBFGS, l1 ratio = 0–1. This approach was chosen as expected to give a better estimation of generalizability, with the algorithm being trained in 4 data folds, and then tested in the remaining fold ensuring the class balance. The Brier score and a calibration curve were calculated for the model on the test set, to assess prediction and calibration loss of predicted probability and lesion class. The ML analysis was performed using the scikit-learn Python package. Accuracy metrics were computed with the same Python package and the caret R package.” [2] (from the article by Romeo et al.; licensed under CC BY 4.0)
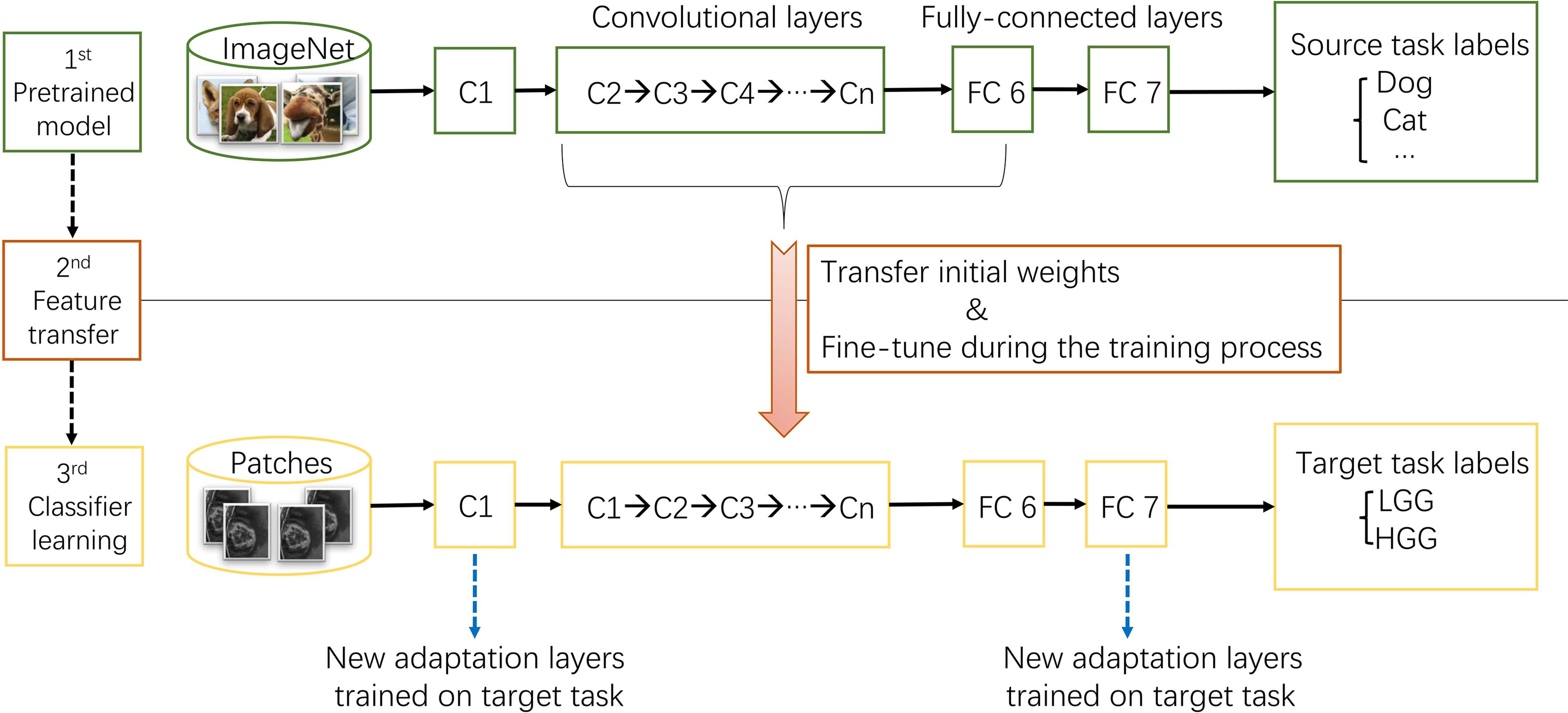
Example#2. “This procedure was explained in Figure […] Thus, the first convolutional layer and fully-connected layers were randomized initialized and freshly trained, in order to accommodate the new object categories. The initial weights of other layers of the pre-trained model were transferred to the new model directly and fine-tuned during the training process for the new task.” [3] (from the article by Yang et al.; licensed under CC BY 4.0)
Figure “The procedure of transfer learning. First, models trained on ImageNet dataset were downloaded. Second, most parameters of the pre-trained models were transferred to the new target task model. Third, first convolutional layer and fully-connected layers were trained on target task.” (from the article by Yang et al.; licensed under CC BY 4.0)
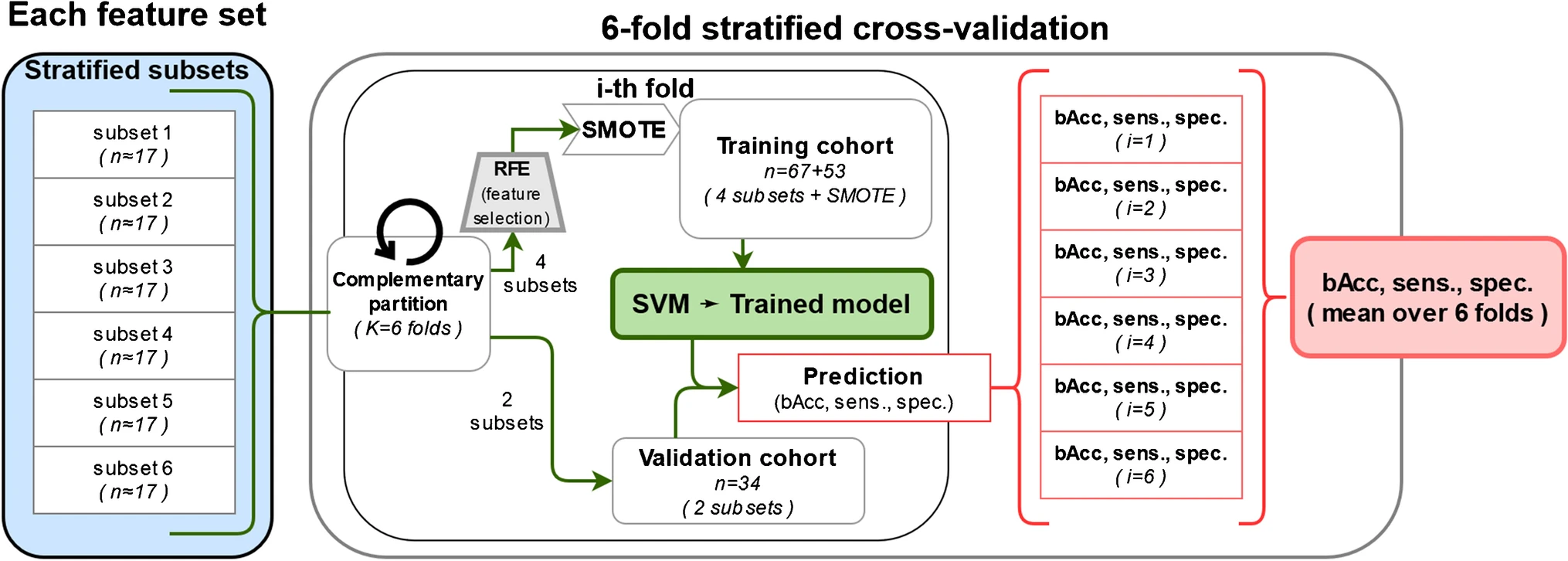
Example#3. “Given the strong class imbalance (59% of the patients belong to the class GGG 1–3), a method to balance the training datasets is required. There are several alternatives, which usually imply an oversampling of the less prevalent classes, an undersampling of the most prevalent class, or a combination of both. With a limited dataset, our study is better suited for oversampling techniques, to avoid neglecting important information. The synthetic minority oversampling technique (SMOTE) was applied to oversample all training features in both less prevalent classes (GGG 4 and GGG 5) up to a 1:1 proportion with the most prevalent class (GGG 1–3). The augmented training data was used to train the SVM for each of the 6 cross-validation cycles. The trained models were then tested in the prediction of GS with the non-augmented validation data. The implemented training and validation of the models is displayed in Fig […]” [4] (from the article by Solari et al.; licensed under CC BY 4.0)
Fig “Training and validation workflow of each SVM model for the prediction of GS” (from the article by Solari et al.; licensed under CC BY 4.0)
Explanation and elaboration of CLEAR item#35
The performance of machine learning models is intrinsically tied to their hyperparameters [5]. A thorough understanding of the tuning process is necessary for assessing the robustness and quality of a study [6]. It is imperative to provide detailed information regarding the training process, also in the supplementary of a study. This includes specifics such as the augmentation technique employed, the stopping criteria for training, the strategy for hyperparameter tuning (e.g., random, grid search, Bayesian), the range of hyperparameter values considered, optimization techniques, regularization parameters, and the initialization of model parameters (e.g., random or transfer learning). If transfer learning is utilized, it is important to clearly state which layers or parameters are frozen or fine-tuned. It is also important to strengthen the reporting with illustrations as demonstrated in Example#2 and Example#3.
References
- Kocak B, Baessler B, Bakas S, et al (2023) CheckList for EvaluAtion of Radiomics research (CLEAR): a step-by-step reporting guideline for authors and reviewers endorsed by ESR and EuSoMII. Insights Imaging 14:75. https://doi.org/10.1186/s13244-023-01415-8
- Romeo V, Cuocolo R, Sanduzzi L, et al (2023) MRI Radiomics and Machine Learning for the Prediction of Oncotype Dx Recurrence Score in Invasive Breast Cancer. Cancers 15:1840. https://doi.org/10.3390/cancers15061840
- Yang Y, Yan L-F, Zhang X, et al (2018) Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front Neurosci 12:804. https://doi.org/10.3389/fnins.2018.00804
- Solari EL, Gafita A, Schachoff S, et al (2022) The added value of PSMA PET/MR radiomics for prostate cancer staging. Eur J Nucl Med Mol Imaging 49:527–538. https://doi.org/10.1007/s00259-021-05430-z
- Leger S, Zwanenburg A, Pilz K, et al (2017) A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep 7:13206. https://doi.org/10.1038/s41598-017-13448-3
- Hatt M, Krizsan AK, Rahmim A, et al (2023) Joint EANM/SNMMI guideline on radiomics in nuclear medicine : Jointly supported by the EANM Physics Committee and the SNMMI Physics, Instrumentation and Data Sciences Council. Eur J Nucl Med Mol Imaging 50:352–375. https://doi.org/10.1007/s00259-022-06001-6