Condition #5
“Does the study include end-to-end deep learning?” [1] (licensed under CC BY)
Explanation
“End-to-end deep learning refers to the use of deep learning to directly process the image data and produce a classification or regression model.” [1] (licensed under CC BY)
Examples from the literature meeting “Yes” condition
Example #1: “Design The models were trained to classify whether an LVO was present. Each case was classified as normal, ICA, M1, or M2. Next, the post-thrombectomy videos were given as input and were reclassified. Movement between classes (either from proximal to distal occlusion or to normal) was deemed as identification of a successful thrombectomy. Finally, the model was used to classify the post-thrombectomy videos as having an mTICI of 3 (complete antegrade reperfusion) or < 3 (incomplete antegrade reperfusion).
Deep learning models
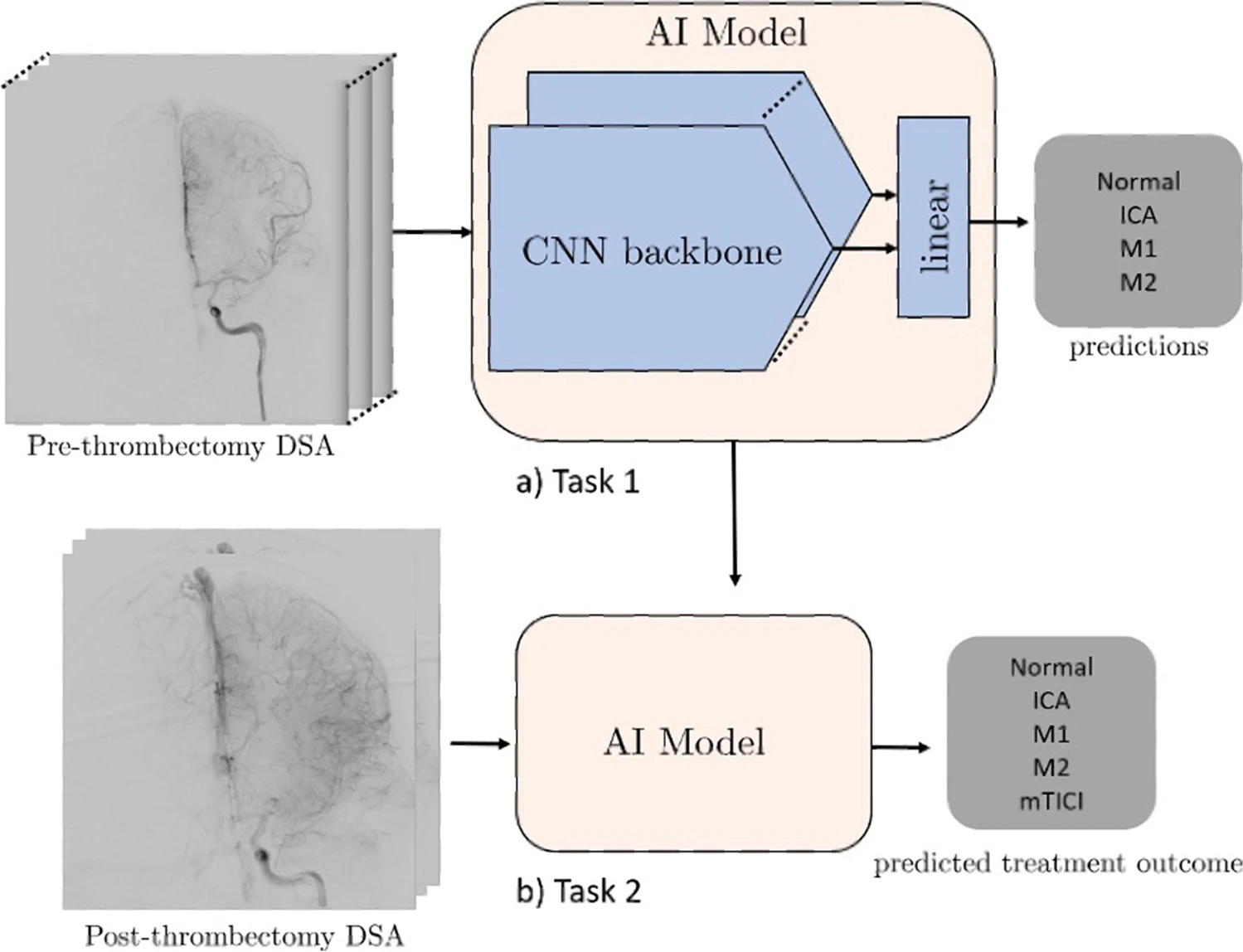
We investigated the performance of several deep learning models using different architectures. First, we employed a 2D convolutional neural network (CNN) model (Xception) which uses only single frames, stacked 2D CNN (stacked-Xception) using multiple frames (2.5D), a 2D vision transformer (ViT), and a 3D CNN (Inception 3D) capturing full special and temporal resolution. On all deep learning approaches except the stacked-Xception + ViT ensemble, we train for 20 epochs with Adam optimizer a learning rate decay of 0.9 every epoch. The ViT component requires considerably longer training times to achieve convergence. For example, training with 20 epochs and 0.9 learning rate decay every epoch results in a training-set F1 score of approximately 0.6 compared to > 0.75 of other models. Data augmentations included horizontal flipping of the frames (for the 2D approaches), cropping, and random brightness distortions. The Xception backbone was pretrained on ImageNet, the inception-3D on Kinetics, and ViT (2D) backbone on Cifar-100. A summary is seen in [Figure 1]. All model architectures are detailed in the supplemental GitHub link (https://github.com/edhlee/DeepMovement).” [2] (licensed under CC BY)
Figure 1. “Summary of DEEP MOVEMENT. Our models are trained and evaluated on DSAs of patients before thrombectomy in task 1 (a). The stacked-Xception model is shown as an example. The pre-thrombectomy model is next evaluated to predict treatment outcome of post thrombectomy to assess the quality of reperfusion in task 2 (b)” [2] (licensed under CC BY)
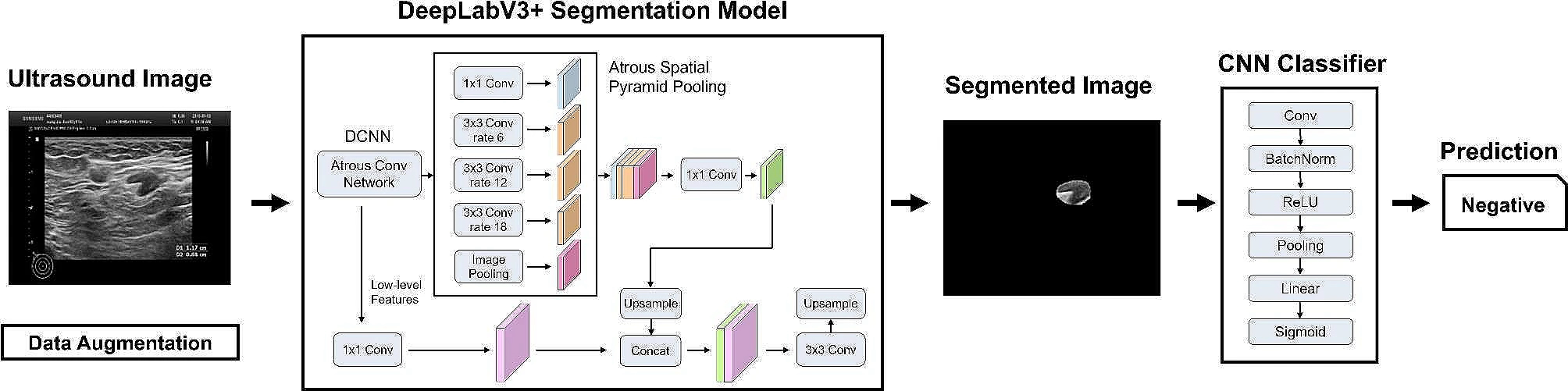
Example #2: “Each patient was labeled according to pathology, as 0 (no metastasis) or 1 (more than one lymph node metastasis). As such, the problem of LN metastasis prediction was addressed as a binary classification problem. The AI system consisted of two pipeline stages, as depicted in [Figure 2]. The first stage utilizes a DeepLabV3 + segmentation model with ResNet-101 as the backbone network that performs pixel-wise localization of the LN over the input ultrasound images. DeepLabV3 + is well-suited for this purpose as it employs atrous convolutions, also known as dilated convolutions, which allow us to capture multi-scale information effectively. This is crucial for accurately detecting lymph nodes of various sizes and shapes in ultrasound images. Subsequently, a simple Convolutional Neural Network (CNN) is implemented to perform the metastasis classification based on the segmented images.Pi∑i=01Pi=1 0<Pi<1. Depending on the clinical application, network parameters of the two stages are optimized together with a loss-weighted backpropagation to enhance the end-to-end performance of the models.” [3] (licensed under CC BY)
Figure 2. “Construction principle of AI” [3] (licensed under CC BY)
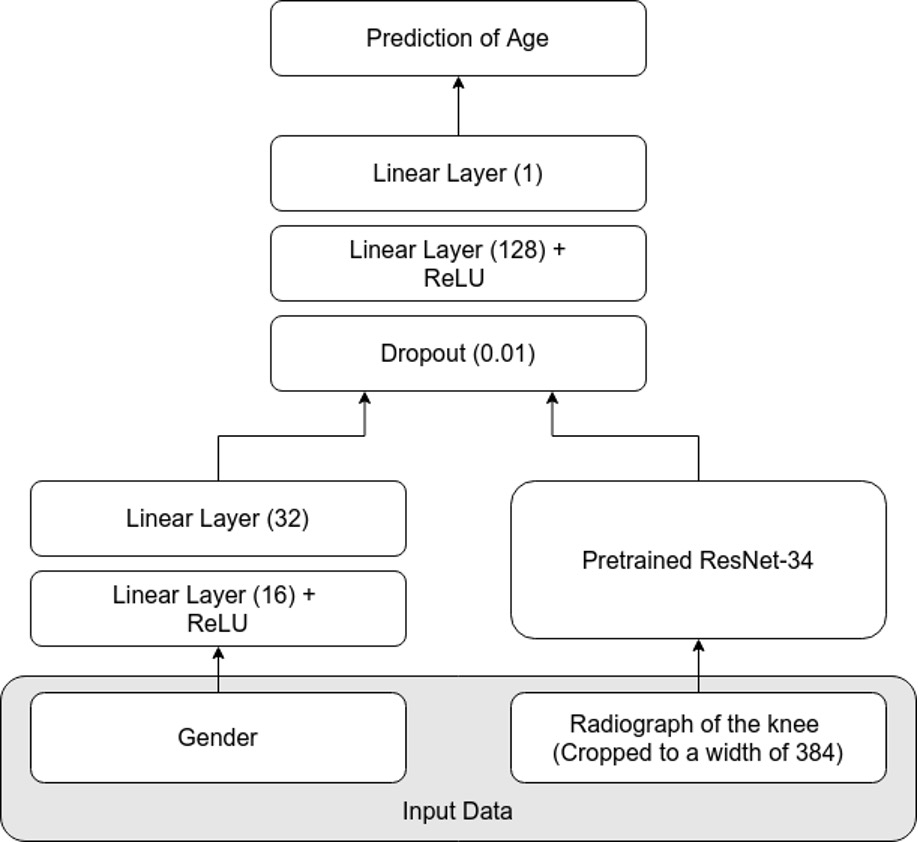
Example #3: “A standard network architecture, the ResNet-34, was used for modeling . Because it is well known that sex has a large impact on the maturity of bones, sex was added as a feature to the network. The ResNet-34 was pretrained on the ImageNet dataset and optimized using the L1 loss and the Adam optimizer. During training, several augmentations were used, which regularizes the network and helps its ability to generalize. The batch size was set to 32. Early stopping was employed to avoid overfitting. Details on the network can be found in Annex 2 [Figure 3].” [4] (licensed under CC BY)
Figure 3. “The network architecture for prediction of chronological age.” [4] (licensed under CC BY)
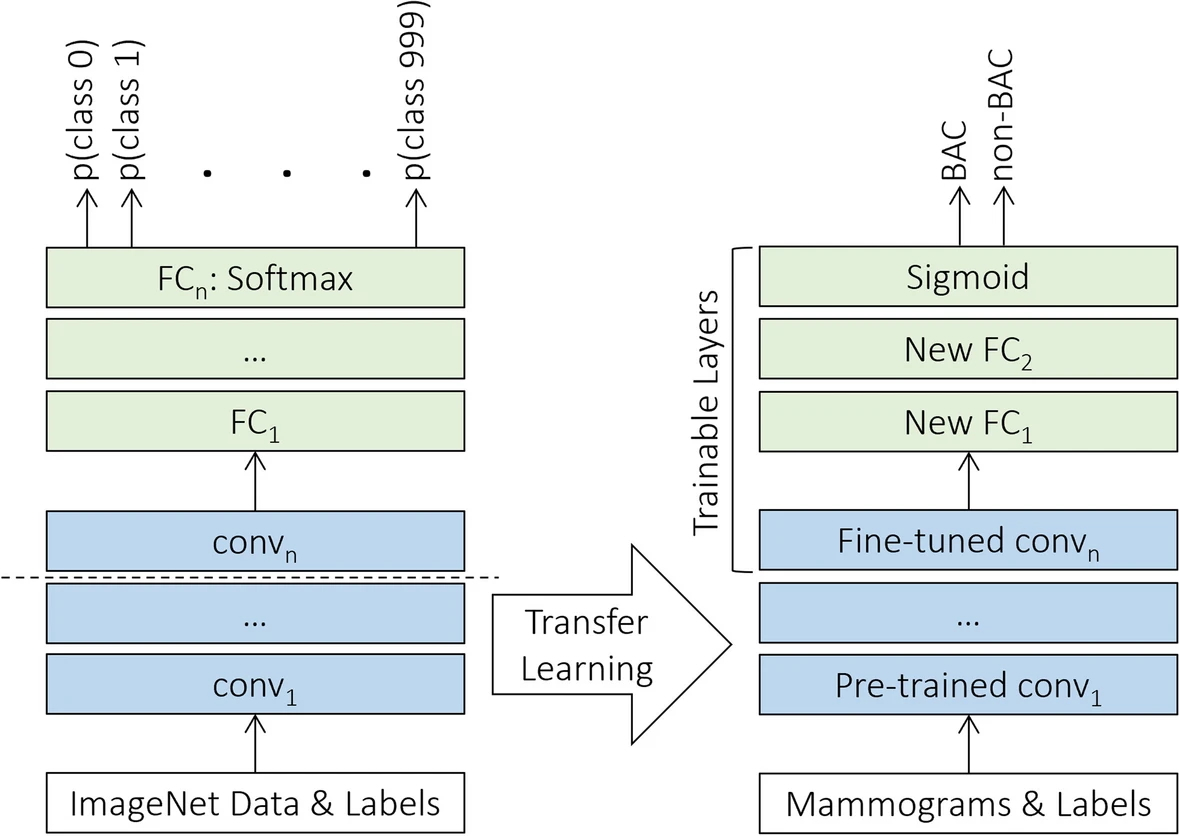
Example #4: “Throughout the experiment, we used a total of eleven deep neural networks, namely Xception , VGG16, VGG19 , ResNet50V2, ResNet101V2, ResNet152V2 , MobileNet , MobileNetV2 , DenseNet121, DenseNet169, and DenseNet201 . The models were previously pretrained on the ImageNet dataset, comprising more than 14 million annotated color images from 1,000 categories , and were publicly available through Keras Applications. Then, we implemented a uniform transfer learning strategy and a harmonized set of hyperparameters across all the networks to directly compare the performance of the various architectures, regardless of specific optimization. Since the source and our target datasets were from disparate domains, the classification layer of each was replaced with two randomly initialized fully connected layers followed by a sigmoid activation function in the output layer, as appropriate for the binary BAC classification task. For transferring knowledge, all layers in the convolutional base except the last were kept frozen with initial pretrained weights, while the rest of the deeper layers and the new classification top were fine-tuned on the mammographic dataset specifically, as illustrated in [Figure 4].” [5] (licensed under CC BY)
Figure 4. “The transfer learning strategy using fine-tuning. FC Fully connected”. [5] (licensed under CC BY)
Hypothetical examples meeting “No” condition
Example #5: Handcrafted radiomic features were extracted from the tumor region of interest in MR images using the Pyradiomics library. A support vector machine (SVM) classifier was then trained on the extracted features to differentiate between glioblastoma and low-grade glioma.
Example #6: A pretrained ResNet-50 model was used to extract deep features from chest X-ray images. These features were then used as input for a logistic regression classifier to predict pneumonia presence.
Importance of the condition
Deep learning (DL) can operate in an end-to-end manner, where input directly produces output without intermediary steps such as manual feature extraction or image pre-processing [6, 7]. End-to-end DL can be integrated partially into study pipelines as components or fully streamlined as standalone systems. In contrast to traditional radiomics or deep feature extraction methods, where deep features are typically subjected to further processing (e.g., feature selection algorithms) and then traditional machine learning (ML) models, end-to-end DL approaches aim to enhance real-world applicability by eliminating complex and time-consuming intermediary steps. However, these approaches are not always superior in predictive performance and remain susceptible to biases and pitfalls [8, 9].
Specifics about the examples meeting “Yes” condition
In Example #1, DL is used to analyze medical videos and determine treatment outcomes. The process incorporates end-to-end DL components, where DL models learn directly from raw video data without manual processing, although it is not entirely clear whether it is end-to-end as a system because different classification schemes are in play after training.
In Example #2, the process involves two stages: segmentation (detecting lymph nodes) and classification (determining metastasis). While both stages employ end-to-end DL models (DeepLabV3+ for segmentation and CNN for classification), the overall system can be considered modular rather than fully end-to-end. Each task is distinct but jointly optimized within the two-stage pipeline, demonstrating end-to-end optimization within each task.
Both Example #1 and Example #2 should be considered positively to meet the METRICS condition due to the end-to-end DL components, though they may not be considered entirely end-to-end DL systems.
In contrast, Example #3 clearly uses a fully end-to-end DL approach using a single ResNet-34 model. This model processes input data and outputs results in one continuous pipeline without separate tasks or models. Similarly, in Example #4, the use of a single DL model qualifies as end-to-end. Here, mammographic images are processed directly to produce classification results, with no manual intervention or external feature extraction. Even with frozen layers or transfer learning, the system remains end-to-end because it connects input to output in one unified training process.
Specifics about the examples meeting “No” condition
In Example #5, radiomic features were manually extracted from MRI images before classification with an SVM. This disrupts the end-to-end process because the deep learning model does not directly learn from the raw images; instead, manual feature extraction is required before classification.
In Example #6, a ResNet-50 model extracted features from chest X-rays, but a logistic regression model handled classification. Since the final classification is handled by a traditional machine learning algorithm rather than a deep learning model that processes the images directly, this breaks the continuity of an end-to-end deep learning approach.
Recommendations for appropriate scoring
To determine whether a study includes end-to-end deep learning (DL) components and meets the condition as “Yes”, it should be verified if a DL model processes input data (e.g., images) directly to generate predictions or classifications without manual feature extraction, meaning it learns directly from input to output.
While most studies employ end-to-end DL components for tasks like classification or segmentation, the overall system may still be modular, comprising separate stages or models. Evaluators should focus on the presence of any end-to-end DL component rather than requiring the entire system to be fully end-to-end.
Special attention is needed for fused models that combine handcrafted and deep radiomics features. Since extracting deep features disrupts the unified input-to-output process, such approaches should be classified as tabular methods rather than end-to-end DL and should be classified as “No”.
Finally, textual descriptions should be assessed alongside figures to accurately determine whether a study incorporates end-to-end DL components.
References
- Kocak B, Akinci D’Antonoli T, Mercaldo N, et al (2024) METhodological RadiomICs Score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 15:8. https://doi.org/10.1186/s13244-023-01572-w
- Kelly B, Martinez M, Do H, et al (2023) DEEP MOVEMENT: Deep learning of movie files for management of endovascular thrombectomy. Eur Radiol 33:5728–5739. https://doi.org/10.1007/s00330-023-09478-3
- Wang X, Nie L, Zhu Q, et al (2024) Artificial intelligence assisted ultrasound for the non-invasive prediction of axillary lymph node metastasis in breast cancer. BMC Cancer 24:910. https://doi.org/10.1186/s12885-024-12619-6
- Demircioğlu A, Quinsten AS, Forsting M, et al (2022) Pediatric age estimation from radiographs of the knee using deep learning. Eur Radiol 32:4813–4822. https://doi.org/10.1007/s00330-022-08582-0
- Mobini N, Capra D, Colarieti A, et al (2024) Deep transfer learning for detection of breast arterial calcifications on mammograms: a comparative study. Eur Radiol Exp 8:80. https://doi.org/10.1186/s41747-024-00478-6
- Castiglioni I, Rundo L, Codari M, et al (2021) AI applications to medical images: From machine learning to deep learning. Physica Medica 83:9–24. https://doi.org/10.1016/j.ejmp.2021.02.006
- Anaya-Isaza A, Mera-Jiménez L, Zequera-Diaz M (2021) An overview of deep learning in medical imaging. Informatics in Medicine Unlocked 26:100723. https://doi.org/10.1016/j.imu.2021.100723
- Lones MA (2024) How to avoid machine learning pitfalls: a guide for academic researchers. Patterns 5:101046. https://doi.org/10.1016/j.patter.2024.101046
- Demircioğlu A (2023) Are deep models in radiomics performing better than generic models? A systematic review. Eur Radiol Exp 7:11. https://doi.org/10.1186/s41747-023-00325-0