Condition #1
“Does the study include segmentation?” [1] (licensed under CC BY)
Explanation
“Segmentation refers to i) fine delineation of a region or volume of interest; ii) rough delineation with bounding boxes; or, iii) cropping the image around a region of interest.” [1] (licensed under CC BY)
Examples from the literature meeting “Yes” condition
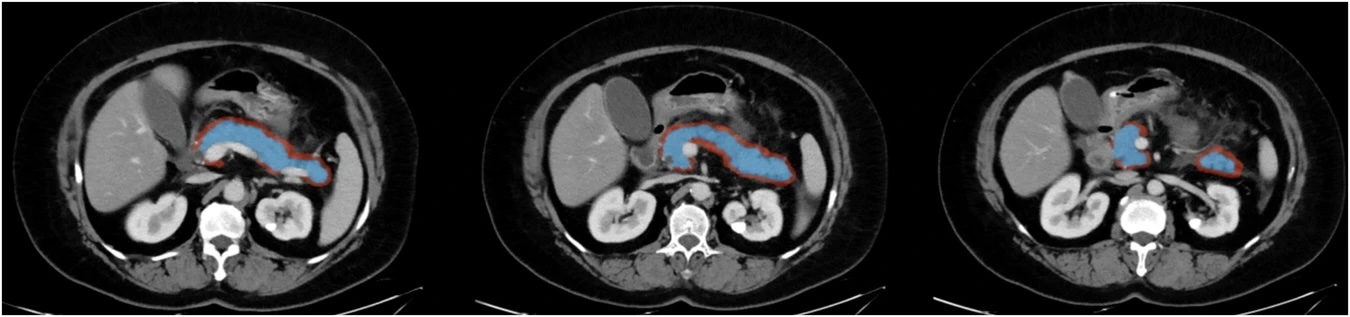
Example #1: “The pancreatic ROIs were manually delineated by the two radiologists on each axial slice along the edge of the pancreatic parenchyma covering the whole pancreatic region. The corresponding peripancreatic ROIs were delineated by expanding the pancreatic ROI by 5 mm towards the peripancreatic area (the peripancreatic area encompassed a 5 mm distance from the pancreatic surface, excluding the pancreatic parenchyma area, blood vessels, bile ducts, peripancreatic lymph nodes and organs).” [2] (licensed under CC BY)
Also see Figure 1.
Figure 1. “Example of regions of interest (ROIs) for acute pancreatitis. The pancreatic ROIs (blue area) were manually delineated along the edge of the pancreatic parenchyma, and the peripancreatic ROIs (red area) were delineated by expanding the pancreatic ROIs by 5 mm towards the peripancreatic area.” [2] (licensed under CC BY)
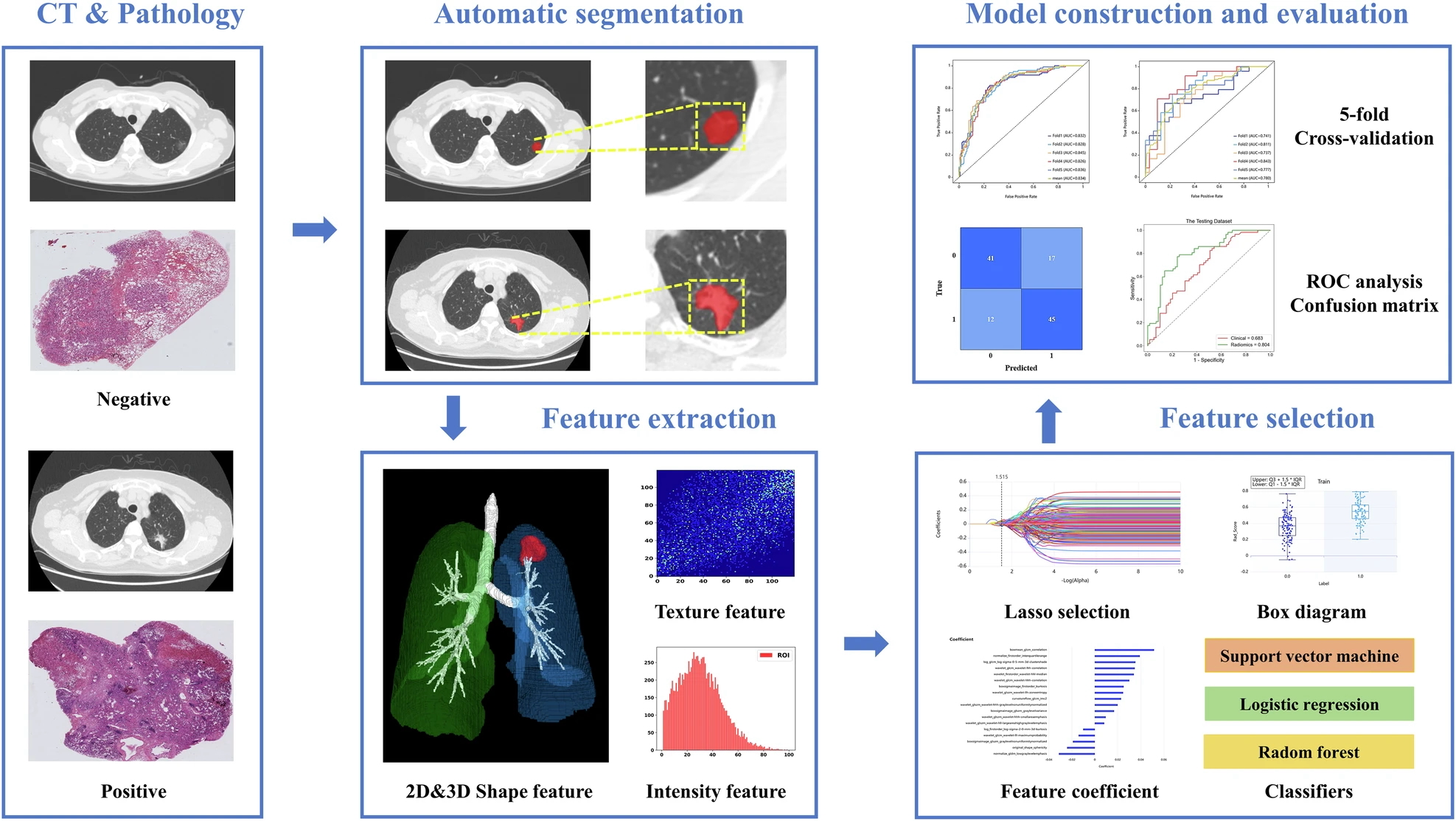
Example #2: “The workflow of radiomics consisted of image segmentation, feature extraction, feature selection, model construction, and evaluation […] A deep learning segmentation model was built with VB-Net to automatically segment lesions. VB-Net is a modified 3D convolutional neural network that combines V-Net with bottleneck structures and is much faster than V-Net.” [3] (licensed under CC BY)
Also see Figure 2.
Figure 2. “The radiomics workflow in the prediction of TLSs. CT, computed tomography; ROC, receiver operating characteristic.” [3] (licensed under CC BY)
Example #3: “To create a cropped deep-learning model for the region of interest (ROI) of a residual-enhancing lesion, a dedicated breast radiologist with 11 years of experience annotated the ROIs on the fifth dynamic subtraction images. Three rectangular-shaped ROIs were placed at the three points (beginning, middle, and ending) of a residual enhancing lesion, fitting to the lesion. […] These ROIs could encompass not only the target lesion but also a small area of surrounding tissue due to its shape. Engineers then utilized these three boxes to generate a 3D voxel ROI, which served as input for the deep-learning models.” [4] (licensed under CC BY)
Hypothetical examples meeting “No” condition
Example #4: A deep-learning model was developed to classify lung cancer subtypes using chest CT images. The entire CT scan was processed through a convolutional neural network (CNN) trained to differentiate between adenocarcinoma, squamous cell carcinoma, and small-cell lung cancer. The model utilized a whole-image approach (i.e., labeling class at the image, exam, or patient level, rather than a region within an image/exam), analyzing global textural and morphological characteristics without explicit segmentation of tumor regions. Feature maps from deep network layers were analyzed to identify relevant patterns contributing to classification.
Example #5: This study investigated osteoporosis classification using lumbar spine DXA images. A machine-learning model was trained on features extracted from the entire image without manually or automatically segmenting vertebral bodies or specific bone regions.
Example #6: A deep-learning model was trained on mammograms to classify breast density categories.
Importance of the condition
Segmentation is a key factor in evaluating the methodological quality of radiomics studies, as it defines the region of interest (ROI) for feature extraction, model training, and clinical interpretation [5]. However, it is not mandatory for all radiomics studies, making segmentation-related items conditional. Its relevance varies depending on the study type and objectives. Evaluators should carefully assess whether segmentation is explicitly reported and adequately described, as it directly impacts the applicability of specific METRICS criteria, hence the final score.
Segmentation can be performed by manual annotation, semi-automatic techniques, or fully automated deep learning-based approaches. It can also be performed semantically (i.e., marking all pixels belonging to a class within an image) or with bounding boxes (which can be used to crop out the region of interest). Each method has distinct methodological implications—manual or automated fine delineation ensures precise ROI definition [6], particularly for lesions or tumors, while bounding boxes and cropping optimize computational efficiency, particularly in deep learning approaches, while maintaining relevant regions [7].
Specifics about the examples meeting “Yes” condition
Example #1 demonstrates a manual segmentation process, with detailed explanations for achieving the final segmentation (e.g., expansion). Example #2 highlights a deep learning-based automatic segmentation as part of the radiomics pipeline. Example #3 uses cropped images focusing on residual enhancing lesions to optimize deep-learning model performance while reducing noise and computational demands.
Specifics about the examples meeting “No” condition
Examples #4 and #5 utilize entire images in their pipeline without performing manual or automatic segmentation of a specific region of interest. In Example #6, no specific region is mentioned, implying that the entire image is used.
Recommendations for appropriate scoring
A study should be classified as “Yes” if segmentation is explicitly mentioned and clearly described. Acceptable methods include manual delineation, semi-automatic segmentation, and automatic segmentation using deep learning, including bounding box/cropping approaches. Evaluators should consider both textual descriptions and visual evidence provided in the paper to ensure accuracy.
A study should be classified as “No” if segmentation is not mentioned, if the study uses a whole-image approach without ROI delineation, or if the description is too vague to confirm whether segmentation was performed.
References
- Kocak B, Akinci D’Antonoli T, Mercaldo N, et al (2024) METhodological RadiomICs Score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 15:8. https://doi.org/10.1186/s13244-023-01572-w
- Chen H, Wen Y, Li X, et al (2025) Integrating CT-based radiomics and clinical features to better predict the prognosis of acute pancreatitis. Insights into Imaging 16:8. https://doi.org/10.1186/s13244-024-01887-2
- Yu Y, Yang T, Ma P, et al (2025) Determining the status of tertiary lymphoid structures in invasive pulmonary adenocarcinoma based on chest CT radiomic features. Insights into Imaging 16:28. https://doi.org/10.1186/s13244-025-01906-w
- Kim S-Y, Lee J, Cho N, Kim Y-G (2024) Deep-learning based discrimination of pathologic complete response using MRI in HER2-positive and triple-negative breast cancer. Sci Rep 14:23065. https://doi.org/10.1038/s41598-024-74276-w
- Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö (2019) Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 25:485–495. https://doi.org/10.5152/dir.2019.19321
- van Timmeren JE, Cester D, Tanadini-Lang S, et al (2020) Radiomics in medical imaging—“how-to” guide and critical reflection. Insights into Imaging 11:91. https://doi.org/10.1186/s13244-020-00887-2
- Lemay A, Gros C, Zhuo Z, et al (2021) Automatic multiclass intramedullary spinal cord tumor segmentation on MRI with deep learning. NeuroImage: Clinical 31:102766. https://doi.org/10.1016/j.nicl.2021.102766