Condition #3
“Does the study include hand-crafted feature extraction?” [1] (licensed under CC BY)
Explanation
“Hand-crafted radiomic features (i.e., traditional radiomic features) are created in advance by human experts or mathematicians” [1] (licensed under CC BY)
Examples from the literature meeting “Yes” condition
Example #1: “Python version 3.12.1 (https://www.python.org) with PyRadiomics package version 3.0.1 (https://pyradiomics.readthedocs.io/en/latest/) was used to extract 18 first-order features and 75 texture features, namely 24 gray-level co-occurrence matrix (GLCM), 14 gray-level run length matrix (GLRLM), 16 gray-level zone length matrix (GLZLM), 16 gray-level dependence matrix (GLDM), and 5 neighborhood gray-tone difference matrix (NGTDM) features” [2] (licensed under CC BY)
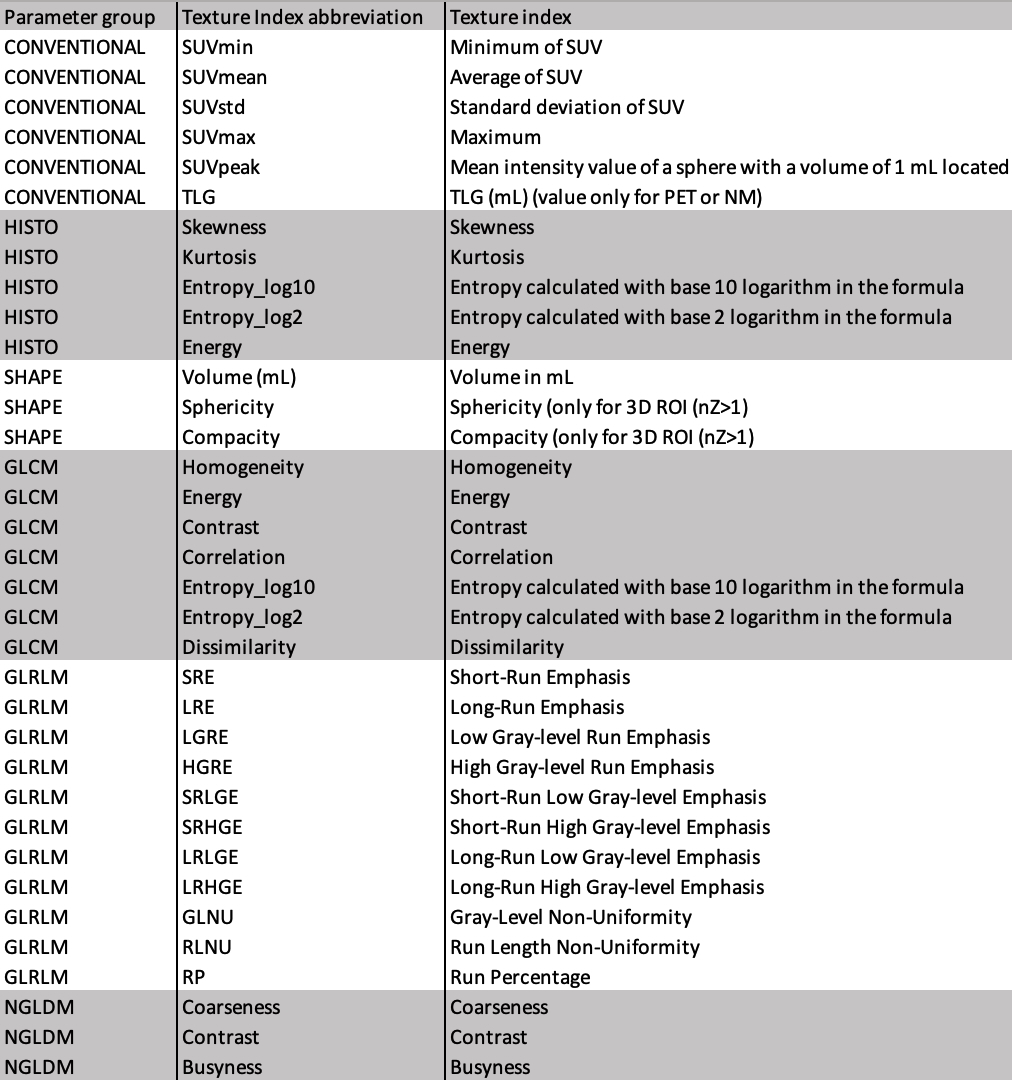
Example #2: “On the whole, 46 parameters [Table 1] were analyzed and computed using the LifeX software”. [3] (licensed under CC BY)
Table 1. “List of radiomics indices.” [3] (cropped; licensed under CC BY)
Example #3: “A total of 59 radiomics features were considered, including 24 derived from the gray-level co-occurrence matrix (GLCM) with a voxel displacement of 1, 16 from the gray-level run length matrix (GLRLM) without distance weighting, 5 from the neighborhood gray-tone difference matrix (NGTDM) with a neighborhood size of 3 × 3 × 3, and 14 from the gray-level dependence matrix (GLDM) with a dependence threshold of 0 and a neighbor distance of 1. Features based on GLCM are designed to capture the spatial relationships between voxel pairs by analyzing how often pairs of voxels with specific intensity values occur in a given spatial configuration. These features focus on local texture patterns and provide insights into the spatial distribution of gray levels, helping to characterize the heterogeneity of the image content at a finer, localized level.” [4] (licensed under CC BY)
Hypothetical examples meeting “No” condition
Example #4: A deep learning-based model was developed using a convolutional neural network (CNN) to extract imaging biomarkers directly from MR images. The model was trained on a dataset of 1,200 scans and employed multiple layers, including convolutional, pooling, and fully connected layers, to learn hierarchical feature representations. The extracted features were then used for classification using a support vector machine (SVM).
Example #5: A self-supervised learning approach was applied to extract radiomics-like features from PET/CT images using a Vision Transformer (ViT) architecture. The model was pre-trained on a large dataset of oncological imaging studies using contrastive learning, enabling it to capture informative imaging features without predefined feature engineering. The extracted features were subsequently fine-tuned for tumor classification and prognosis prediction.
Importance of the condition
Radiomics features can be either handcrafted or deep. Handcrafted features are extracted using a certain series of predefined mathematical equations, whereas deep radiomics features are extracted from one or more layers of a neural network, such as a convolutional neural network [5]. Advantages of handcrafted radiomics are their standardized nature [6], whereas deep radiomics are considered to be more robust [7].
Specifics about the examples meeting “Yes” condition
Example #1 presents the extraction of handcrafted radiomics features using the standardized method of PyRadiomics, a widely used Python package for radiomics feature extraction. PyRadiomics is compliant with the Image Biomarker Standardisation Initiative (IBSI), ensuring that its feature definitions adhere to established guidelines for reproducibility and standardization. While PyRadiomics primarily functions as a Python package, it can also be integrated into other software platforms, such as 3D Slicer (slicer.org). Example #2 demonstrates the extraction of handcrafted features using LIFEx, another well-established software tool that adheres to IBSI guidelines. Both PyRadiomics and LIFEx are among the recognized IBSI-compliant software implementations, a list of which can be found at https://theibsi.github.io/implementations/. Example #3 describes a set of extracted radiomics features without specifying a software package or an explicit reference to IBSI compliance. Despite this, these features are still classified as handcrafted, but the absence of a standardized extraction method may impact their reproducibility and comparability across studies.
Specifics about the examples meeting “No” condition
Examples #4 and #5 clearly rely on deep learning-based feature extraction rather than predefined, handcrafted radiomics features, making them negative examples for Condition #3.
Recommendations for appropriate scoring
A paper should be classified as “Yes” if it explicitly provides classes or formulas for predefined radiomics features (e.g., GLCM), regardless of the tool or standardization method used. However, the use of tools like PyRadiomics or standardized methods such as IBSI can indicate the usage of handcrafted features.
Evaluators should be cautious of deep learning-based features and should classify such studies as “No”.
For studies employing both handcrafted and deep learning-based features as separate pipelines, evaluators may choose to assess these pipelines separately and apply the METRICS independently, resulting in two distinct scores for the handcrafted (Condition = Yes) and deep learning-based pipelines (Condition = No). However, a single METRICS evaluation can be used to represent the overall study quality (Condition = Yes), accounting for the need for such a study to fulfill the requirements applying to all the different techniques employed.
References
- Kocak B, Akinci D’Antonoli T, Mercaldo N, et al (2024) METhodological RadiomICs Score (METRICS): a quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 15:8. https://doi.org/10.1186/s13244-023-01572-w
- Zhu L, Dong H, Sun J, et al (2025) Robustness of radiomics among photon-counting detector CT and dual-energy CT systems: a texture phantom study. Eur Radiol 35:871–884. https://doi.org/10.1007/s00330-024-10976-1
- Kallos-Balogh P, Vas NF, Toth Z, et al (2024) Multicentric study on the reproducibility and robustness of PET-based radiomics features with a realistic activity painting phantom. PLOS ONE 19:e0309540. https://doi.org/10.1371/journal.pone.0309540
- Cepero A, Yang Y, Young L, et al (2024) Longitudinal FDG-PET Radiomics for Early Prediction of Treatment Response to Chemoradiation in Locally Advanced Cervical Cancer: A Pilot Study. Cancers 16:3813. https://doi.org/10.3390/cancers16223813
- Kim S, Kim BR, Chae H-D, et al (2022) Deep Radiomics-based Approach to the Diagnosis of Osteoporosis Using Hip Radiographs. Radiol Artif Intell 4:e210212. https://doi.org/10.1148/ryai.210212
- Shur JD, Doran SJ, Kumar S, et al (2021) Radiomics in Oncology: A Practical Guide. Radiographics 41:1717–1732. https://doi.org/10.1148/rg.2021210037
- Vrettos K, Triantafyllou M, Marias K, et al (2024) Artificial intelligence-driven radiomics: developing valuable radiomics signatures with the use of artificial intelligence. BJR Artificial Intelligence 1:ubae011. https://doi.org/10.1093/bjrai/ubae011